Rich, responsive 3D graphics
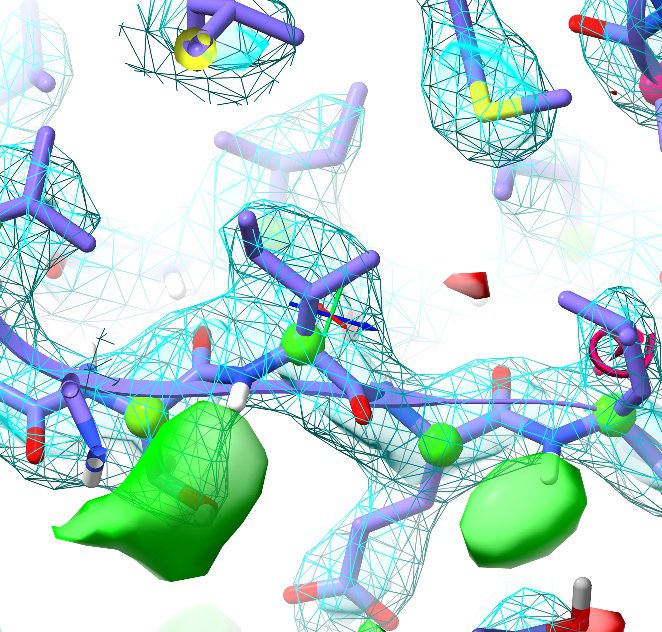
ISOLDE's implementation as a plugin to UCSF ChimeraX ensures that it inherits all the capabilities that come with a dedicated molecular graphics package. Atom graphics, volume surfaces (as wireframe, solid/transparent or any combination of the above) and all markup are rendered at 30-60 frames per second.